rm(list=ls()) PSYC122-w19-how-to
Introduction
In Week 19, we aim to further develop skills in working with the linear model.
We do this to learn how to answer research questions like:
- What person attributes predict success in understanding?
- Can people accurately evaluate whether they correctly understand written health information?
In this class, what is new is our focus on critically evaluating the ways in which people, effects or results can vary using interaction analyses.
- This work simulates the kinds of interaction analyses that psychologists must often do in professional research.
Naming things
I will format dataset names like this:
all.data.csv
I will also format variable (data column) names like this: variable
I will also format value or other data object (e.g. cell value) names like this: all.data
I will format functions and library names like this: e.g. function ggplot() or e.g. library {tidyverse}.
The data we will be using
In this how-to guide, we use data from:
(1.) two 2020 studies of the response of adults from a UK national sample to written health information
study-one-general-participants.csvstudy-two-general-participants.csv
(2.) along with data recently collected from PSYC122 students.
These data have been joined together to create a dataset of over 400 observations. This is because it often requires a lot of evidence to be able to draw precise or accurate conclusions about potential interaction effects.
Answers
Step 1: Set-up
To begin, we set up our environment in R.
Task 1 – Run code to empty the R environment
Task 2 – Run code to load relevant libraries
library("ggeffects")
library("sjPlot")
library("tidyverse")── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.2.0 ✔ readr 2.1.6
✔ forcats 1.0.1 ✔ stringr 1.6.0
✔ ggplot2 4.0.2 ✔ tibble 3.3.1
✔ lubridate 1.9.4 ✔ tidyr 1.3.2
✔ purrr 1.2.1
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
✖ ggplot2::set_theme() masks sjPlot::set_theme()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsStep 2: Load the data
Task 3 – Read in the data file we will be using
The data file is called:
all-data.csv
Use the read_csv() function to read the data file into R:
all.data <- read_csv("all.data.csv") Rows: 478 Columns: 11
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (5): participant_ID, study, GENDER, EDUCATION, ETHNICITY
dbl (6): mean.acc, mean.self, AGE, SHIPLEY, HLVA, FACTOR3
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Task 4 – Inspect the data file
Use the summary() function to take a look at the dataset.
summary(all.data) participant_ID mean.acc mean.self study
Length:478 Min. :0.3600 Min. :2.250 Length:478
Class :character 1st Qu.:0.7500 1st Qu.:6.200 Class :character
Mode :character Median :0.8300 Median :7.200 Mode :character
Mean :0.8066 Mean :6.967
3rd Qu.:0.9000 3rd Qu.:7.910
Max. :1.0000 Max. :9.000
AGE SHIPLEY HLVA FACTOR3
Min. :18.00 Min. :19.00 Min. : 2.000 Min. : 9.00
1st Qu.:21.00 1st Qu.:31.00 1st Qu.: 7.000 1st Qu.:46.00
Median :29.00 Median :35.00 Median : 9.000 Median :50.00
Mean :32.85 Mean :34.19 Mean : 8.929 Mean :49.84
3rd Qu.:43.00 3rd Qu.:38.00 3rd Qu.:10.000 3rd Qu.:55.00
Max. :76.00 Max. :40.00 Max. :14.000 Max. :63.00
GENDER EDUCATION ETHNICITY
Length:478 Length:478 Length:478
Class :character Class :character Class :character
Mode :character Mode :character Mode :character
Notice that we conducted a series of studies using the same online survey methods.
- Our data include the responses made by PSYC122 students.
Step 3: Work with different kinds of variables
It is an important skill in psychological data analysis to be able to:
(1.) identify the different kinds of data variables we are working with; (2.) adapt our methods depending on differences in kind (variable Class or type).
We learn about these skills, next, in exercises that combine some revision with some new moves.
Revise: consolidate what you know
Task 5 – Use summaries or plots to examine variables
Q.1. What is the mean of the
AGEvariable in the dataset?A.1. You can use the
summary()function to calculate that the meanAGEis 32.85Q.2. Draw a histogram of the age distribution.
A.2. You can write the code as you have been shown to do previously (e.g., in week 16):
ggplot(data = all.data, aes(x = AGE)) +
geom_histogram(binwidth = 1) +
theme_bw() +
labs(x = "Age (years)", y = "frequency count")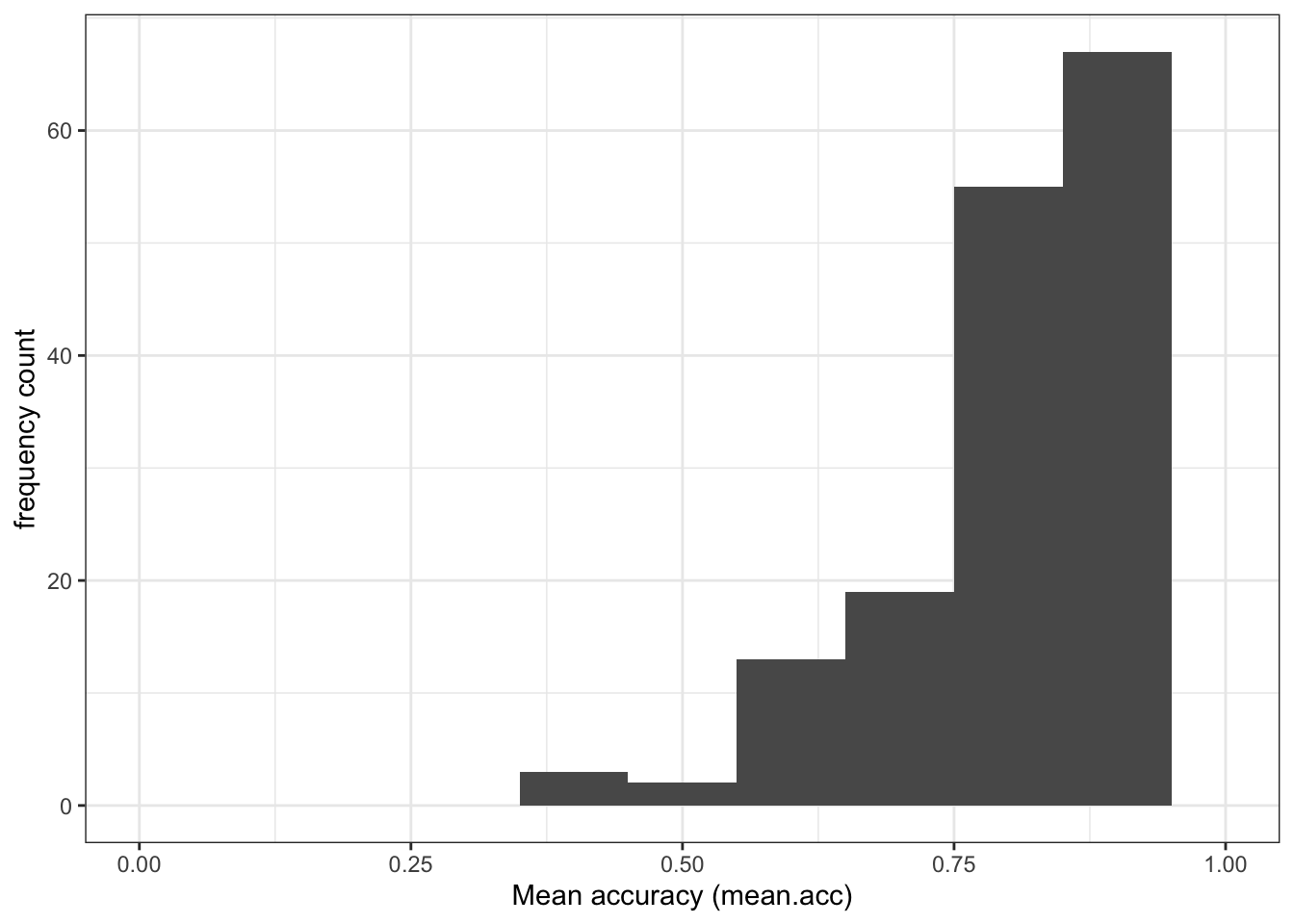
- Q.3. Examine the age distribution: what does it tell you about the ages of the participants in the sample?
- A.3. The histogram shows that most participants are around 20 years in age but there is a “long tail” of small numbers of older participants.
Extend: make some new moves
We can tell from the summary and the distribution that the AGE variable consists of numbers (participant ages in years).
Another way to identify what kind of variable we have is by using a check question:
is.factor(all.data$AGE)[1] FALSEis.numeric(all.data$AGE)[1] TRUEThe is._() family of functions act like identity checks: what is this variable? These functions work like questions with TRUE or FALSE answers.
- Is this variable a factor? [TRUE or FALSE]
- Is this variable numeric? [TRUE or FALSE]
Revise: consolidate what you know
Task 6 – Identify (and change) what kind of variable – numeric or factor – a variable is
Hint: Task 6 – The is._() functions are available to ask what kind a variable is
Hint: Task 6 – The as._() functions can be used to change variable types
- Q.4. What kind of variable is
EDUCATION? - A.4. Use the same functions but change the variable name:
is.factor(all.data$EDUCATION)[1] FALSEis.numeric(all.data$EDUCATION)[1] FALSEQ.5. What do the
is._()functions tell us?A.5. The function calls tell us that
EDUCATIONis not a factor and is not numeric.Q.6. If you look at the
summary()results, what does it tell you theClassofEDUCATIONis?A.6. The summary shows that
EDUCATIONis identified asClass: character.
You can check this by doing the is._() test again.
is.character(all.data$EDUCATION)[1] TRUE- Q.7. How do you change the class or type of the variable
EDUCATION? Change the variable’s type (Class) into a factor. - A.7. The
as._()functions can be used to change variable type, as you have done previously (e.g., in week 16).
all.data$EDUCATION <- as.factor(all.data$EDUCATION)- Q.8. What does a
summary()tell us aboutEDUCATION?
summary(all.data) participant_ID mean.acc mean.self study
Length:478 Min. :0.3600 Min. :2.250 Length:478
Class :character 1st Qu.:0.7500 1st Qu.:6.200 Class :character
Mode :character Median :0.8300 Median :7.200 Mode :character
Mean :0.8066 Mean :6.967
3rd Qu.:0.9000 3rd Qu.:7.910
Max. :1.0000 Max. :9.000
AGE SHIPLEY HLVA FACTOR3
Min. :18.00 Min. :19.00 Min. : 2.000 Min. : 9.00
1st Qu.:21.00 1st Qu.:31.00 1st Qu.: 7.000 1st Qu.:46.00
Median :29.00 Median :35.00 Median : 9.000 Median :50.00
Mean :32.85 Mean :34.19 Mean : 8.929 Mean :49.84
3rd Qu.:43.00 3rd Qu.:38.00 3rd Qu.:10.000 3rd Qu.:55.00
Max. :76.00 Max. :40.00 Max. :14.000 Max. :63.00
GENDER EDUCATION ETHNICITY
Length:478 Further:224 Length:478
Class :character Higher :254 Class :character
Mode :character Mode :character
- A.8. The summary will show how many different participants report themselves as belonging to one of the different
EDUCATIONlevel groups in the corresponding survey question.
You should see counts of the numbers of people in different EDUCATION level groups in the total survey sample:
EDUCATION Further:224 Higher :254
We can see that 224 participants reported in the survey that they had attained a Further level of education (corresponding to A-level or Sixth Form).
254 participants reported a Higher level of education (corresponding to university level).
Step 4: Now draw plots to examine associations between variables and to visualize interactions
Here, we are going to build on previous work to use plots to:
(1.) Examine the associations between pairs of variables, an outcome and a predictor (2.) Examine how the association between two variables can be different for different levels of a third variable
Scenario (2.) comes up if we are thinking about or working with interaction effects.
Consolidation: practice to strengthen skills
Task 7 – Create a scatterplot to examine the association between two numeric variables
Hint: Task 7 – We are working with geom_point() and you need x and y aesthetic mappings.
Run a chunk of code to make the plot.
ggplot(data = all.data, aes(x = HLVA, y = mean.acc)) +
geom_point() +
theme_bw() +
labs(y = "Accuracy of understanding (mean.acc)",
x = "Health literacy (HLVA)") +
xlim(0, 16) + ylim(0, 1)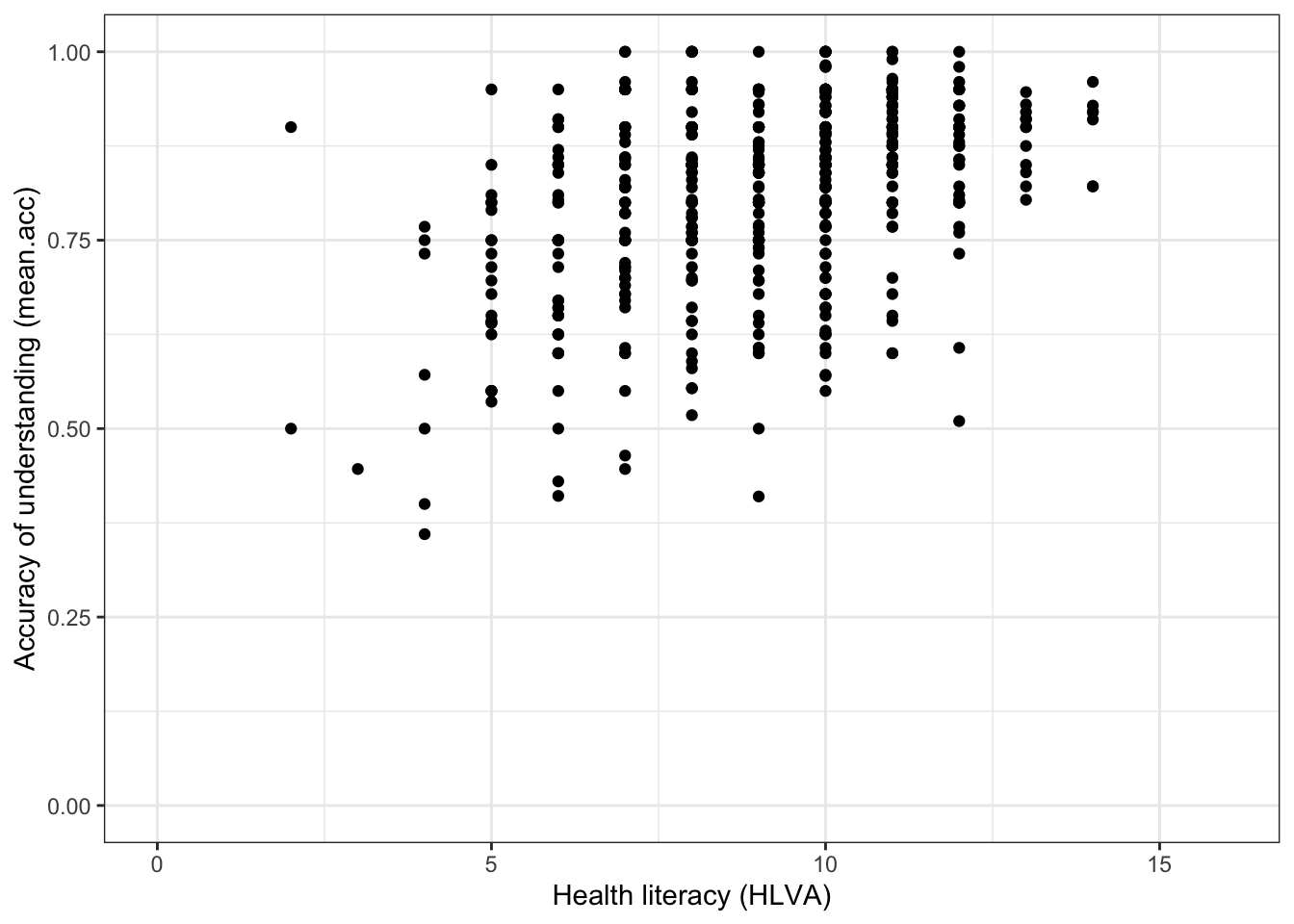
This plot shows:
- the possible association between x-axis variable
HLVAand y-axis variablemean.acc.
The plot code moves through the following steps:
ggplot(...)make a plot;ggplot(data = all.data, ...)with theall.datadataset;ggplot(...aes(x = HLVA, y = mean.acc))using two aesthetic mappings
x = HLVAmapHLVAvalues to x-axis (horizontal, left to right) positions;y = mean.accmapmean.accvalues to y-axis (vertical, bottom to top) positions;
geom_point()show the mappings as points.theme_bw()changes the theme to a white background.labs(y = "Accuracy of understanding (mean.acc)", x = "Health literacy (HLVA)")changes axis labels.xlim(0, 6) + ylim(0, 1)changes axis limits.
Questions: Task 7
- Q.9. What do you notice about the distribution of
mean.accscores at increasing values of health literacy (HLVA) scores? - A.9. The scatterplot shows that, for increasing health literacy (
HLVA) scores, there is an associated rise in accuracy of understanding (mean.acc) scores.
Introduce: make some new moves
In the next sequence of exercises, we are going to take the all.data dataset and split it into parts (sub-sets) in different ways.
- We split the dataset vertically, into different sets of rows or observations.
Why are we learning how to do this?
The capacity to do this kind of split-analyse operation is extremely useful.
We are going to do this so that we can examine how the effects of interest to us (here, associations) can vary between different groups of people.
Remember, observations about different people are on different rows in
all.data.We are going to focus on an association between two column variables (
mean.acc,HLVA).We are going to identify different groups (sub-sets) of observations using values on a third variable.
Task 8 – Create a scatterplot to examine the association between two numeric variables, showing how the association is different for different values of a third variable
Hint: Task 8 – We are working with geom_point() again.
Hint: Task 8 – We can show how the association between two variables (mean.acc, HLVA) is different for different values of a third, categorical (factor), variable (EDUCATION) by splitting the plot so that we see the association for different levels of EDUCATION.
Here, we are splitting the dataset into:
- observation data from people with just
EDUCATIONlevelFurther - observation data from people with just
EDUCATIONlevelHigher
and we are then producing a different scatterplot for each sub-set of data produced by the split.
Run the following chunk of code to make the plot.
ggplot(data = all.data, aes(x = HLVA, y = mean.acc)) +
geom_point() +
geom_smooth(method = 'lm') +
theme_bw() +
labs(y = "Accuracy of understanding (mean.acc)",
x = "Health literacy (HLVA)") +
xlim(0, 16) + ylim(0, 1) +
facet_wrap(~ EDUCATION)`geom_smooth()` using formula = 'y ~ x'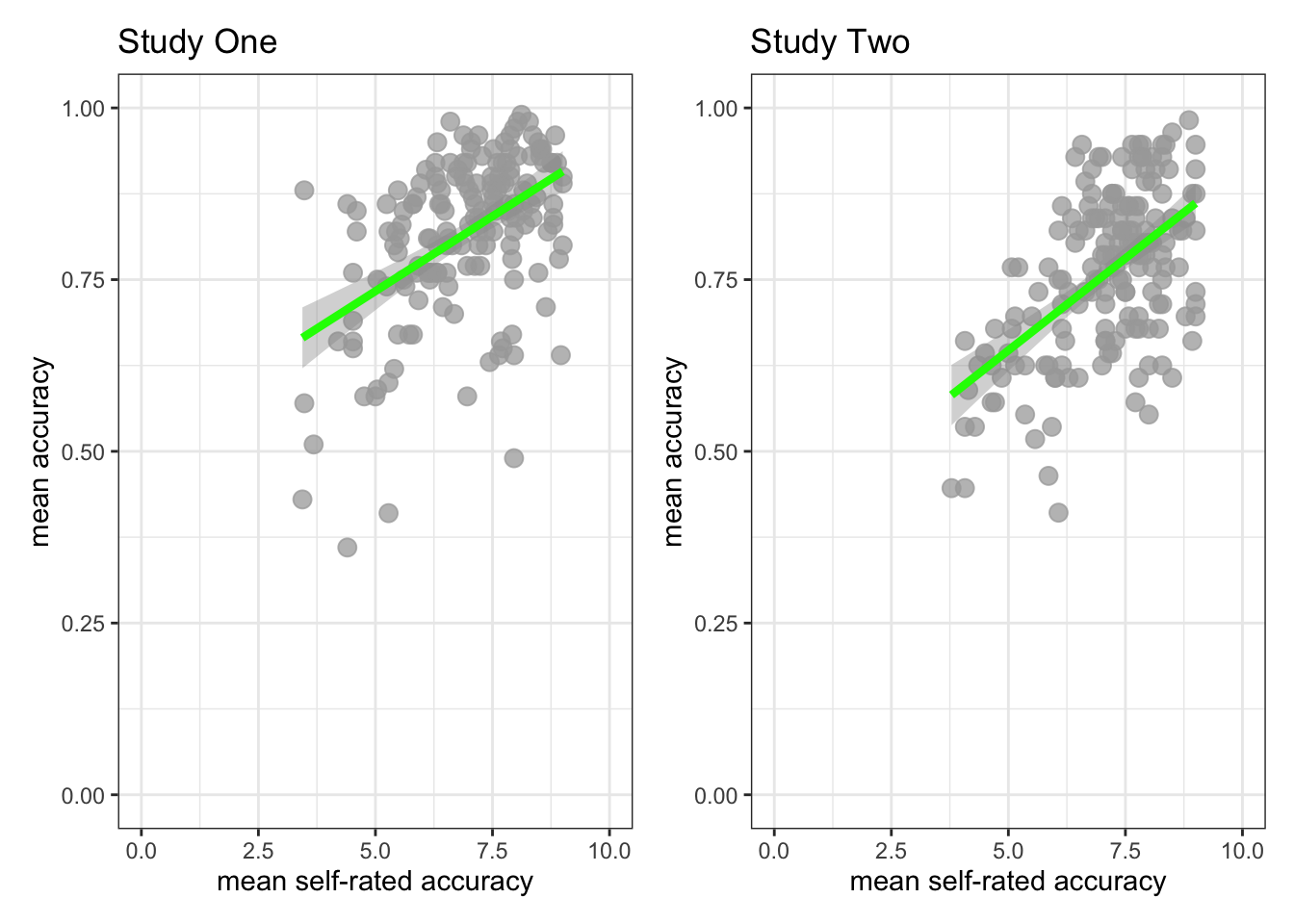
This plot shows:
- the possible association between x-axis variable
HLVAand y-axis variablemean.acc - for different groups of people – people with
Furtheror people withHigherlevels ofEDUCATION.
The plot code moves through steps we have seen before. What is new here is this bit:
facet_wrap(~ EDUCATION)
The function facet_wrap() splits the dataset into sub-sets (different parts): different sets of rows, where different sets are defined according to different values of the variable identified inside the brackets (~ EDUCATION).
Here, we are asking for different scatterplots for the dataset split by whether participants are coded as people with Further or people with Higher levels of EDUCATION.
You can see a general guide to the function here:
Questions: Task 8
- Q.10. What do you notice about the distribution of
mean.accscores at increasing values of health literacy (HLVA) scores, for different sub-sets of the data: for people withFurthercompared to people withHigherlevels ofEDUCATION? - A.10. You can see that the association between
mean.accandHLVAscores appears to be similar for different levels of education: the distribution of points is different but the shape is similar overall, and the trend line representing the association betweenmean.accandHLVAlooks similar too.
Hint: Task 8 – We can show how the association between two variables (mean.acc, HLVA) is different for different values of a third numeric variable (SHIPLEY) by splitting the dataset
Here, we are first creating a new variable to code for (distinguish between) different sub-sets of observations (different rows in the dataset) based on SHIPLEY scores, and then second drawing different plots based on different subsets.
We do this in two steps, as follows.
First, run a chunk of code to divide (cut) the dataset into parts:
all.data$SHIPLEY_splits <- cut_number(all.data$SHIPLEY, 3)The code works bit-by-bit like this:
all.data$SHIPLEY_splits <-create a new variable,SHIPLEY_splitsand add it to the datasetall.datagiven the work done by the bit of code on the right of the arrow<-.
On the right of the arrow <- …
cut_number(all.data$SHIPLEY, 3)uses thecut_number(...)to divide the dataset observations into three sets based on the values in theall.data$SHIPLEYvariable named inside the brackets.
The function works by ordering all the observations (the rows) in the dataset according to the values in the named variable all.data$SHIPLEY, then splitting the dataset (here, into three sub-sets) based on values in that named variable.
- Given this code, we are going to get three sub-sets of the data, observations corresponding to (1.) low (2.) mid or (3.) high
SHIPLEYvalues. - We split the data into data about people with:
(1.) low SHIPLEY values, scores between 19-33 on the SHIPLEY test; (2.) mid SHIPLEY values, scores between 33-37 on the SHIPLEY test; (3.) high SHIPLEY values, scores between 37-40 on the SHIPLEY test.
You can see a guide to the function here:
https://ggplot2.tidyverse.org/reference/cut_interval.html
Note that where we split the data (what ranges of scores we use) will depend on the sample.
Second, we draw a plot to examine if the association between two variables (mean.acc, HLVA) is different for different values of a third variable (SHIPLEY) by splitting the data:
ggplot(data = all.data, aes(x = HLVA, y = mean.acc)) +
geom_point() +
geom_smooth(method = 'lm') +
facet_wrap(~ SHIPLEY_splits, nrow = 1, labeller = label_both) +
labs(y = "Accuracy of understanding (mean.acc)",
x = "Health literacy (HLVA)") +
theme_bw() `geom_smooth()` using formula = 'y ~ x'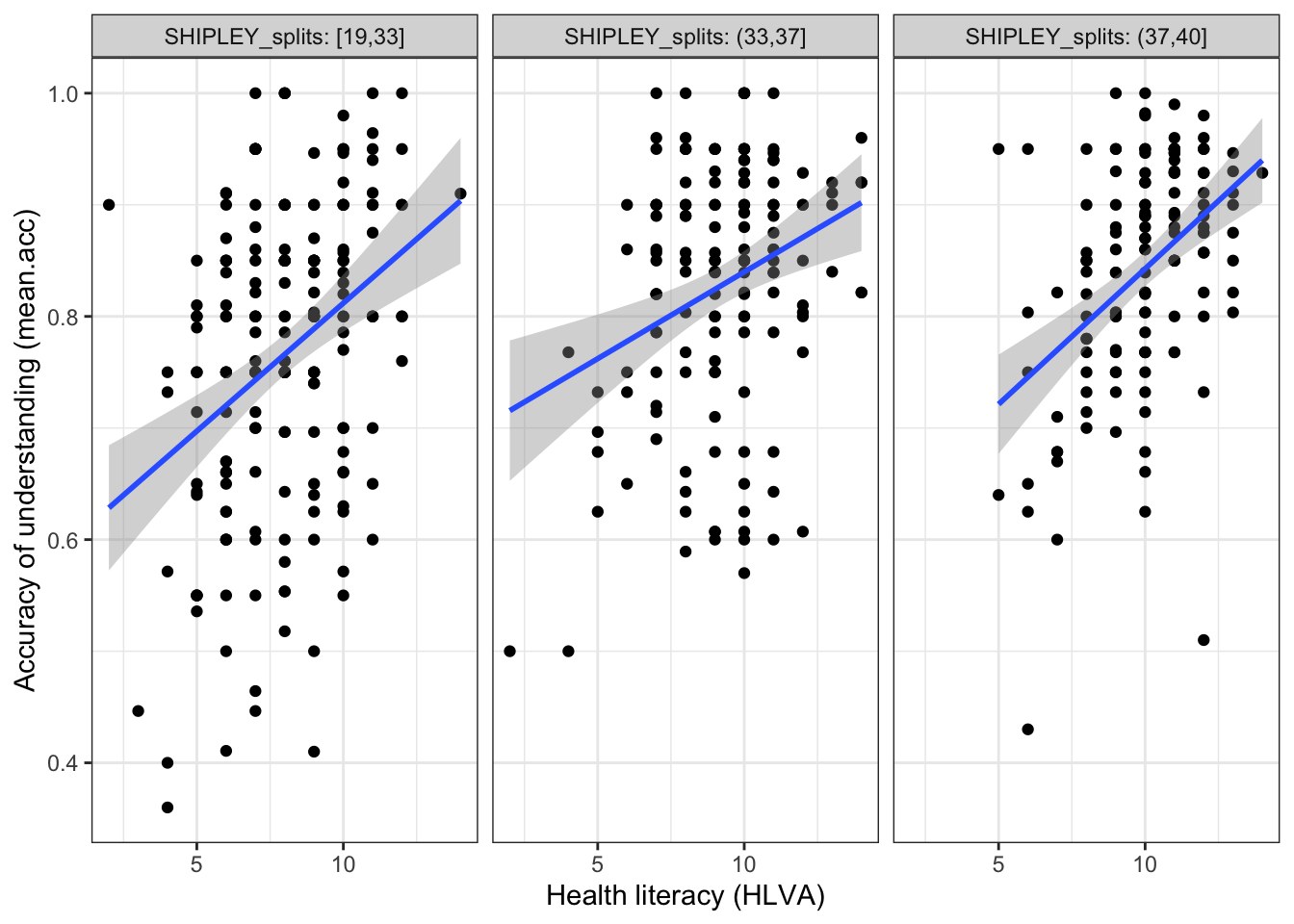
The plot code moves through the scatterplot production steps we have seen before.
The bit that is new is here:
facet_wrap(~ SHIPLEY_splits, nrow = 1, labeller = label_both) which is included to use the data split, constructed earlier.
This addition allows us to show:
- the nature of the association between two variables (
mean.acc,HLVA) - for different values of the third variable (
SHIPLEY) - by splitting the data into three sub-sets according to
SHIPLEYscore (19-33 low, 33-37 mid, 37-40 high).
Questions: Task 8
Q.11. What do you notice about the distribution of
mean.accscores at increasing values of health literacy (HLVA) scores, for different sub-sets of the data: for (1.) low (2.) mid (3.) highSHIPLEYvalues?A.11. You can see that the association between
mean.accandHLVAscores appears to vary (the slope differs) given different values of the third variable (SHIPLEY).Q.12. What do you think the values in the grey labels at the top of the facets (the plot panels) tell us?
A.12. The values tell us what values (e.g.,
19,33) are used to identify the range ofSHIPLEYscores that have been used to define the sub-set of observations for which the plot below shows the association betweenmean.accandHLVAscores.
Step 5: Use linear models with multiple predictors, to estimate the effects of factors as well as numeric variables
Consolidation: practice to strengthen skills
As we saw in weeks 17 and 18, we can use linear models to predict outcome variables.
- Linear models can include numeric variables (like
HLVA) as predictors. - Linear models can also include categorical variables or factors (like
EDUCATION) as predictors.
Task 9 – First, fit a linear model including just numeric variables as predictors
Hint: Task 9 – We use the lm() function to fit a model with mean.acc as the outcome and HLVA and SHIPLEY as predictors.
Note that in this model:
HLVAis a measure of health literacy – numeric scores correspond to a test of knowledge of health-related wordsSHIPLEYis a measure of vocabulary knowledge – numeric scores correspond to a test of knowledge of vocabulary
model <- lm(mean.acc ~ HLVA + SHIPLEY,
data = all.data)
summary(model)
Call:
lm(formula = mean.acc ~ HLVA + SHIPLEY, data = all.data)
Residuals:
Min 1Q Median 3Q Max
-0.38048 -0.06818 0.01317 0.07672 0.30910
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.448687 0.040197 11.162 < 2e-16 ***
HLVA 0.019132 0.002555 7.488 3.43e-13 ***
SHIPLEY 0.005471 0.001252 4.370 1.53e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.1127 on 475 degrees of freedom
Multiple R-squared: 0.2003, Adjusted R-squared: 0.1969
F-statistic: 59.48 on 2 and 475 DF, p-value: < 2.2e-16If you look at the model summary you can answer the following questions.
Q.13. What is the estimate for the coefficient of the effect of the predictor,
SHIPLEY(vocabulary knowledge)?A.13. 0.005471
Q.14. Is the effect significant?
A.14. It is significant. Because
Pr(>|t|)equals1.53e-05for theEstimateof the effect ofSHIPLEY(vocabulary knowledge): this means that p < .05, and so the effect is significant.
Hint: Task 9 – Do you know how to intepret numbers like 1.53e-05? If you are unsure you can find out about scientific notation here:
https://www.calculatorsoup.com/calculators/math/scientific-notation-converter.php
Q.15. What are the values for t and p for the significance test for the estimated coefficient of the effect of
HLVA(health literacy)?A.15. t = 7.488, p = 3.43e-13
Q.16. What do you conclude are the effects of the
SHIPLEY(vocabulary knowledge) andHLVA(health literacy) variables, as predictors of outcomemean.acc(accuracy of understanding of health information)?A.16. The model slope estimates suggests that both predictors are significant, and that as both
SHIPLEY(vocabulary knowledge) andHLVA(health literacy) increase, somean.accscores are predicted to increase also.
Task 10 – Second, fit a linear model including a numeric variable and a factor as predictors
Hint: Task 10 – We use the lm() function to fit a model with mean.acc as the outcome and FACTOR3 and EDUCATION as predictors.
Note that in this model:
FACTOR3is a measure of reading strategy – numeric scores correspond to a test of strategy awarenessEDUCATIONis a measure of education level – factor levels correspond to self-reported education
model <- lm(mean.acc ~ FACTOR3 + EDUCATION,
data = all.data)
summary(model)
Call:
lm(formula = mean.acc ~ FACTOR3 + EDUCATION, data = all.data)
Residuals:
Min 1Q Median 3Q Max
-0.42828 -0.06774 0.02177 0.09198 0.31055
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.6032460 0.0415482 14.519 < 2e-16 ***
FACTOR3 0.0040226 0.0008266 4.866 1.55e-06 ***
EDUCATIONHigher 0.0053666 0.0113014 0.475 0.635
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.1229 on 475 degrees of freedom
Multiple R-squared: 0.04889, Adjusted R-squared: 0.04489
F-statistic: 12.21 on 2 and 475 DF, p-value: 6.75e-06If you look at the model summary you can answer the following questions.
Q.17. What is the estimate for the coefficient of the effect of the predictor,
FACTOR3(strategy)?A.17. 0.0040226
Q.18. Is the effect significant?
A.18. It is significant. Because
Pr(>|t|)equals1.55e-06for theEstimateof the effect ofFACTOR3(strategy): this means that p < .05, and so the effect is significant.Q.19. What are the values for t and p for the significance test for the estimated coefficient of the effect of
EDUCATION(level,Furthercompared toHigher)?A.19. t = .475, p = .635
Q.20. Is the effect of
EDUCATIONsignificant?A.20. It is not significant. Because
Pr(>|t|)equals0.635for theEstimateof the effect ofEDUCATIONlevel this means that p > .05, and so the effect is not significant.
Hint: Task 10 – How do we know how to interpret estimates for the effects of factors? We discussed how R deals with factors in week 18, and discuss factors, and the effects of factors, in the week 19 lecture.
Q.21. How should we interpret the coefficient estimate for the effect of
EDUCATION?A.21. Though the effect is not significant, we can say that, on average, outcome
mean.accis 0.0053666 (slightly) higher for people withHigher(compared toFurther) level of education.Q.22. What do you conclude are the effects of the
FACTOR3(strategy) andEDUCATION(level,Furthercompared toHigher) variables, as predictors of outcomemean.acc(accuracy of understanding of health information)?A.22. The model slope estimates suggest that education level does not have a significant effect but that as
FACTOR3(strategy) scores increase, somean.accscores are predicted to increase.
Step 6: Use linear models with multiple predictors, including interaction effects
Introduce: make some new moves
As we saw through Task 8, it is possible that:
- the association between two variables (
mean.acc,HLVA) can be different for different values of a third categorical (factor) variable (EDUCATION); - the association between two variables (
mean.acc,HLVA) can be different for different values of a third variable (SHIPLEY)
While we can examine these possibilities using visualizations, we often need to conduct statistical analyses to test interaction effects, or the ways in which the association between two variables can be different for different levels of a third variable.
We can extend linear models to include terms that allow us to test or to estimate interaction effects.
Task 11 – Center numeric variables before using them as predictors
Hint: Task 11
When we include numeric variables (e.g., HLVA, SHIPLEY) in linear models, it can cause problems of interpretation or of estimation if we include the variables as they first come to us in datasets (as raw variables).
- It often helps our analyses to center variables on their means.
- Centering a variable means calculating its mean, and then subtracting that mean from every value in the variable column.
For example, to center values of the FACTOR3 variable, we create a new column data$FACTOR3_centered by first calculating the mean of the data$FACTOR3 variable, and then subtracting that mean from every row value in data$FACTOR3.
all.data$FACTOR3_centered <- all.data$FACTOR3 - mean(all.data$FACTOR3, na.rm = TRUE)- Q.23. Can you check what is going on when we center variables?
- Q.23. Hint: You can see what is happening when you run this code by first calculating the mean for
FACTOR3
mean(all.data$FACTOR3, na.rm = TRUE)[1] 49.841then looking at column values in the new FACTOR3_centered column, compared to the original FACTOR3 column:
all.data %>%
select(FACTOR3_centered, FACTOR3) %>%
print(n = 20)# A tibble: 478 × 2
FACTOR3_centered FACTOR3
<dbl> <dbl>
1 10.2 60
2 -3.84 46
3 1.16 51
4 -4.84 45
5 13.2 63
6 6.16 56
7 3.16 53
8 -15.8 34
9 5.16 55
10 10.2 60
11 0.159 50
12 9.16 59
13 11.2 61
14 -9.84 40
15 -6.84 43
16 5.16 55
17 8.16 58
18 -7.84 42
19 -3.84 46
20 -2.84 47
# ℹ 458 more rowsWhat is the difference between the FACTOR3 and FACTOR3_centered columns?
A.23. You can see that every value in the
FACTOR3_centeredcolumn equals the value, on the same row, in theFACTOR3column, minus the mean forFACTOR3.Q.24. Can you center the variable
HLVA?A.24. You can use the same code to center
HLVA:
all.data$HLVA_centered <- all.data$HLVA - mean(all.data$HLVA, na.rm = TRUE)- Q.25. Can you center the variable
SHIPLEY? - A.25. You can use the same code to center
SHIPLEY:
all.data$SHIPLEY_centered <- all.data$SHIPLEY - mean(all.data$SHIPLEY, na.rm = TRUE)Why are we learning how to do this?
It is common in data analysis to want to transform variables before using them in analyses.
- Here, we are transforming predictor variables by centering them on their mean values.
The practical reason for doing this transformation now is because if we use uncentered (raw) numeric variables as predictors in linear models that involve interactions the model can find it difficult to distinguish between the effects of those predictors and the effect of their interaction.
Task 12 – Specify models to include interaction effects: interactions between two numeric predictor variables
Hint: Task 12
We use lm() to specify models, as we have been doing.
- What changes is that we now use the
*operator in our model code to specify interaction effects.
For example, let’s suppose that we want to examine the possibility that the effect of health literacy (HLVA) on accuracy understanding (mean.acc) is different for different values of vocabulary knowledge (SHIPLEY).
- Here, we are considering the possibility that the association between health literacy (
HLVA) and accuracy of understanding (mean.acc) varies. - We are considering the possibility that the the association between health literacy (
HLVA) and accuracy of understanding (mean.acc) is different for different groups of people. - We are considering the possibility that the effect of health literacy (
HLVA) on accuracy of understanding (mean.acc) is different for people who differ, also, in vocabulary knowledge (SHIPLEY).
In examining the possibility that the effect of health literacy (HLVA) on accuracy of understanding (mean.acc) is different for different values of vocabulary knowledge (SHIPLEY), we are examining a possible interaction between the effects of health literacy (HLVA) and vocabulary knowledge (SHIPLEY).
Let’s go through this step-by-step.
First, consider: if we ignored the possibility of an interaction, we would fit a model with just the predictors: health literacy (HLVA) and vocabulary knowledge (SHIPLEY).
We can fit the same model that we fit before (for Task 9).
- We vary the code a little, by using the centered variables we have just created:
model <- lm(mean.acc ~ HLVA_centered + SHIPLEY_centered,
data = all.data)
summary(model)
Call:
lm(formula = mean.acc ~ HLVA_centered + SHIPLEY_centered, data = all.data)
Residuals:
Min 1Q Median 3Q Max
-0.38048 -0.06818 0.01317 0.07672 0.30910
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.806587 0.005155 156.453 < 2e-16 ***
HLVA_centered 0.019132 0.002555 7.488 3.43e-13 ***
SHIPLEY_centered 0.005471 0.001252 4.370 1.53e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.1127 on 475 degrees of freedom
Multiple R-squared: 0.2003, Adjusted R-squared: 0.1969
F-statistic: 59.48 on 2 and 475 DF, p-value: < 2.2e-16Q.26. Can you compare the summary of results for this model with the model, using the same (but not centered) predictors, that you fitted for Task 9 – compare the estimates, what do you see?
A.26. If we compare the results of the models we can see that the estimates, t-test values and probability values are the same for
HLVAcompared toHLVA_centeredor forSHIPLEYcompared toSHIPLEY_centered.This shows that centering predictors only changes the scaling of the predictor variables so that their centers are 0.
Second, now consider: we aim to test or estimate the interactions between the effects of health literacy (HLVA_centered) and vocabulary knowledge (SHIPLEY_centered).
- We vary the code a little, again, this time by separating the predictors by
*not by+:
model <- lm(mean.acc ~ HLVA_centered*SHIPLEY_centered,
data = all.data)
summary(model)
Call:
lm(formula = mean.acc ~ HLVA_centered * SHIPLEY_centered, data = all.data)
Residuals:
Min 1Q Median 3Q Max
-0.37986 -0.06840 0.01342 0.07652 0.29872
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.8061527 0.0054951 146.703 < 2e-16 ***
HLVA_centered 0.0191754 0.0025645 7.477 3.70e-13 ***
SHIPLEY_centered 0.0055241 0.0012742 4.335 1.78e-05 ***
HLVA_centered:SHIPLEY_centered 0.0001132 0.0004922 0.230 0.818
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.1128 on 474 degrees of freedom
Multiple R-squared: 0.2004, Adjusted R-squared: 0.1953
F-statistic: 39.59 on 3 and 474 DF, p-value: < 2.2e-16Q.27. Can you compare the summary of results for this model with the results for the previous model (which uses the same predictors but without an interaction)?
What do you see?
A.27. If we compare the results of the models we can see that now the summary gives us estimates for the effects of:
HLVA_centered– this is the effect ofHLVA_centeredwhenSHIPLEY_centeredis 0SHIPLEY_centered– this is the effect ofSHIPLEY_centeredwhenHLVA_centeredis 0HLVA_centered:SHIPLEY_centered– this is the interaction, and reflects the change in the effect ofHLVA_centeredon outcomemean.acc, given different values ofSHIPLEY_centered.Notice that the effect of the interaction is not significant, suggesting, for this sample, that the effects of these predictor variables are at least approximately independent of each other.
The * symbol (the * operator) is used here to code for a model including the effects of the two variables separated by the symbol, here, as well as the effect of the interaction between these two variables.
Task 13 – Specify models to include interaction effects: interactions between a numeric predictor variable and a factor predictor variable
Hint: Task 13 – We follow the same approach that we followed through Task 12.
Now consider: we aim to test or estimate the interactions between the effects of reading strategy (FACTOR3) and education level (EDUCATION).
- We vary the code to change the predictor variables but the structure otherwise stays the same:
model <- lm(mean.acc ~ FACTOR3_centered*EDUCATION,
data = all.data)
summary(model)
Call:
lm(formula = mean.acc ~ FACTOR3_centered * EDUCATION, data = all.data)
Residuals:
Min 1Q Median 3Q Max
-0.43422 -0.06111 0.01700 0.09208 0.23754
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.802704 0.008193 97.977 <2e-16 ***
FACTOR3_centered 0.002210 0.001100 2.008 0.0452 *
EDUCATIONHigher 0.005249 0.011241 0.467 0.6408
FACTOR3_centered:EDUCATIONHigher 0.004104 0.001656 2.479 0.0135 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.1223 on 474 degrees of freedom
Multiple R-squared: 0.06106, Adjusted R-squared: 0.05512
F-statistic: 10.28 on 3 and 474 DF, p-value: 1.45e-06Q.28. Can you identify what effects are shown in the model summary?
A.28. We can see that now the summary gives us estimates for the effects of:
FACTOR3_centered– this is the effect of reading strategy (FACTOR3) whenEDUCATIONlevel isFurtherEDUCATIONHigher– this is the effect ofEDUCATIONwhenFACTOR3_centeredis 0.
Note that the effect of EDUCATION is the estimated difference in outcome when EDUCATION level is Further (not shown, treated as the baseline) compared to when EDUCATION level is Higher.
FACTOR3_centered:EDUCATIONHigher– this is the interaction, and reflects the change in the effect ofFACTOR3_centeredon outcomemean.acc, given different values ofEDUCATION, here, whenEDUCATIONisHigherinstead ofFurther.
What are we learning here?
Remember from week 18:
Categorical variables or factors and reference levels.
- If you have a categorical variable like
EDUCATIONthen when you use it in an analysis, R will look at the different categories (calledlevels) e.g., here,Higher, Further` and it will pick one level to be the reference or baseline level. - The reference is the the level against which other levels are compared.
- Here, the reference level is
Further(education) simply because, unless you tell R otherwise, it picks the level with a category name that begins earlier in the alphabet as the reference level.
How do we interpret interactions?
- It helps to plot the effects estimates, given the model.
We can do this in a two-step sequence, similar to the sequence we trialled through weeks 17 and 18:
- First fit the model
- Second, use the model information to plot the predictions, given the effects
# -- first fit the model
model <- lm(mean.acc ~ FACTOR3_centered*EDUCATION,
data = all.data)
# -- make plot
plot_model(model, type = "pred",
terms = c("FACTOR3_centered", "EDUCATION")) +
theme_bw() +
ylim(0, 1)Scale for y is already present.
Adding another scale for y, which will replace the existing scale.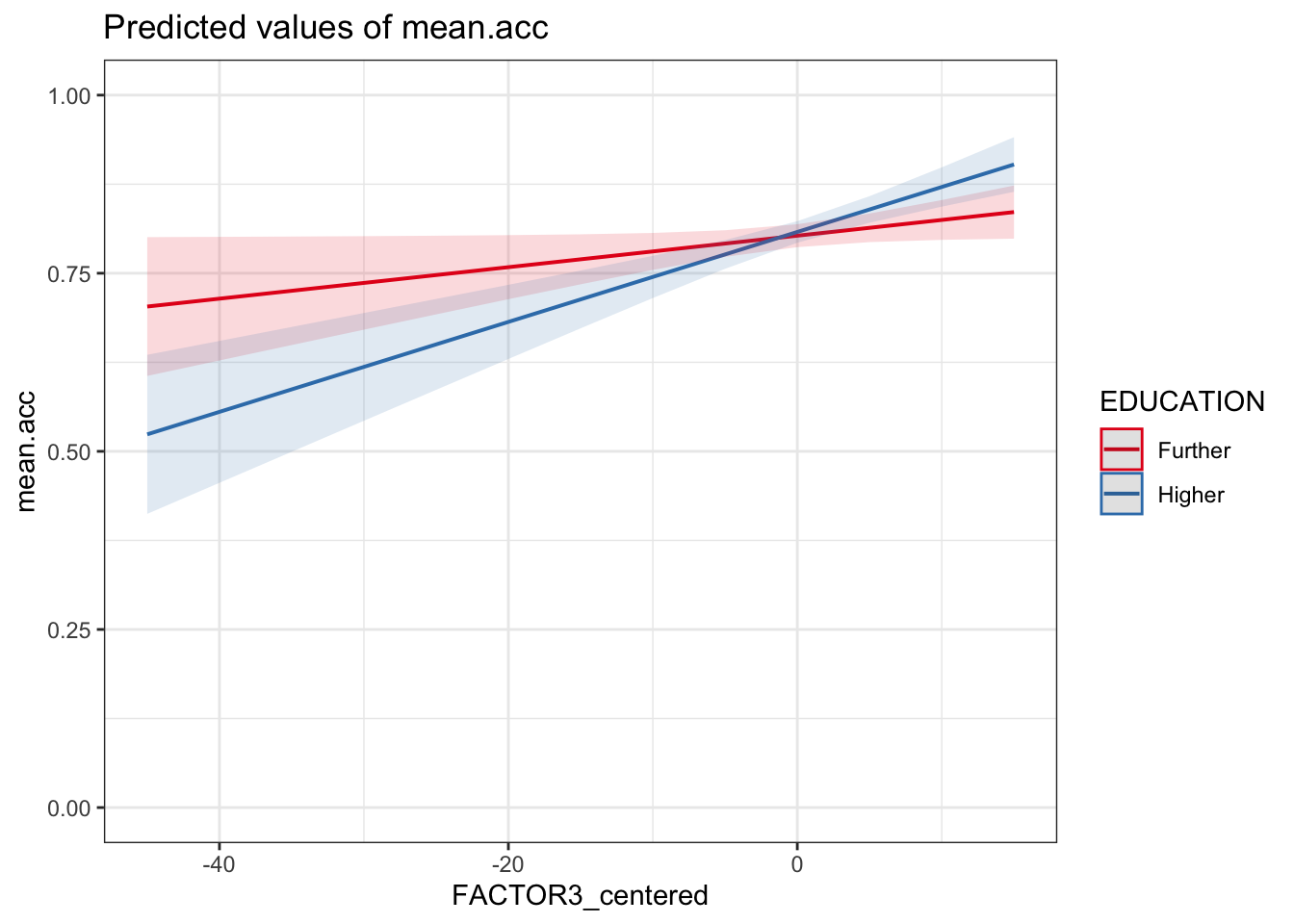
Q.29. Can you identify how the effects of reading strategy (
FACTOR3) and education level (EDUCATION) interact, given the model results summary and this plot?A.29. The plot, and the estimate for the interaction
FACTOR3_centered:EDUCATIONHigher, together show that the coefficient for the effect of reading strategy (FACTOR3_centered) is larger (the slope is steeper) whenEDUCATIONisHigherinstead ofFurther.Notice that, for this sample, the interaction is not significant, suggesting that there is no difference between education levels in the relative effectiveness of reading strategy.